
Wet and in silico
A complete systems biology approach
Experimentations, modelling and simulation
Systems Biology investigates system’s properties such as, e.g., cell size, growth and division. These aspects cannot be derived from the study of single genes or proteins, or genes and proteins networks, but need an understanding of their quantitative relationship, their interaction and their dynamic, all of which necessitate the usage of mathematical models as well as precise experimentation.
SYSBIO published some papers with a complete systems biology approach, like, for instance:
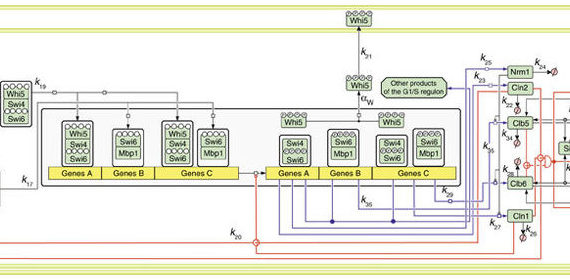
- a mathematical dynamic model of the basic molecular mechanism controlling the G1/S transition in budding yeast (Palumbo et al. (2016) Nat Commun, 7:11372)
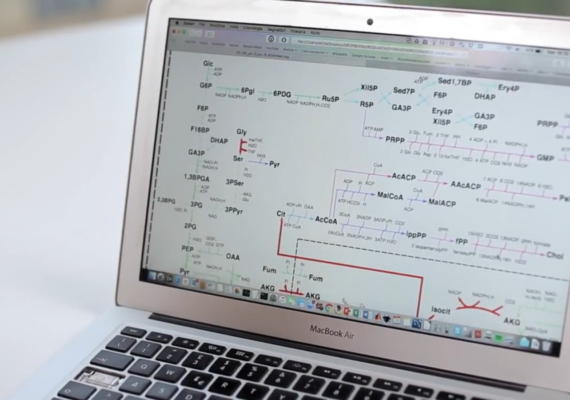
- a metabolic core model (constraint-based) that showed that the increased utilization of glucose and glutamine, with enhanced glutamine-dependent lactate production, promotes cancer cell growth. We call this effect the “the WarburQ effect” (Damiani et al. (2017) PLoS Comput. Biol., 13, 9:e1005758).
More details below.
A complete dynamic model
A dynamic math model of G1/S transition in yeast
Here we present a mathematical model of the basic molecular mechanism controlling the G1/S transition, whose major regulatory feature is multisite phosphorylation of nuclear Whi5.
A contraint-based metabolic model
A metabolic core model showing how glucose, glutamine and lactate promotes cancer cell growth
Flux Balance Analysis
We collect findings that offer new understanding of the logic of the metabolic reprogramming that underlies cancer cell growth: enhanced utilization of glucose and glutamine, with increased glutamine-dependent lactate production, promotes cancer cell growth. We call it the the “WarburQ effect”
Systems Biology is interdisciplinary and typically involves collaborations from the fields of biology, medicine, engineering, information sciences and mathematics, chemistry and physics. The heart of the systems biology approach is an iterative process between laboratory experiments and mathematical modelling, using computers. Based on large volumes of quantitative data at the molecular, cellular, tissue and organ level, algorithms are used to create models which allow predictions to be made of the behaviour of complex biological systems with the aim of gaining an overall understanding of the system
